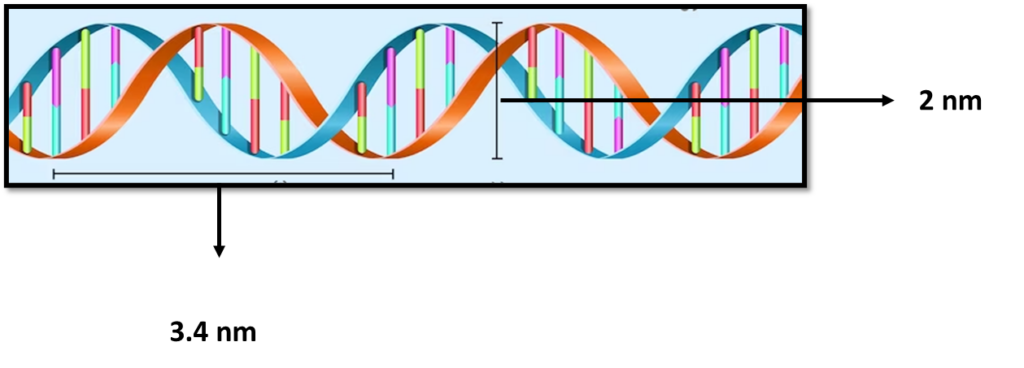
Deoxyribonucleic acid (DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix structure. This polymer carries genetic instructions for all known organisms and many viruses.
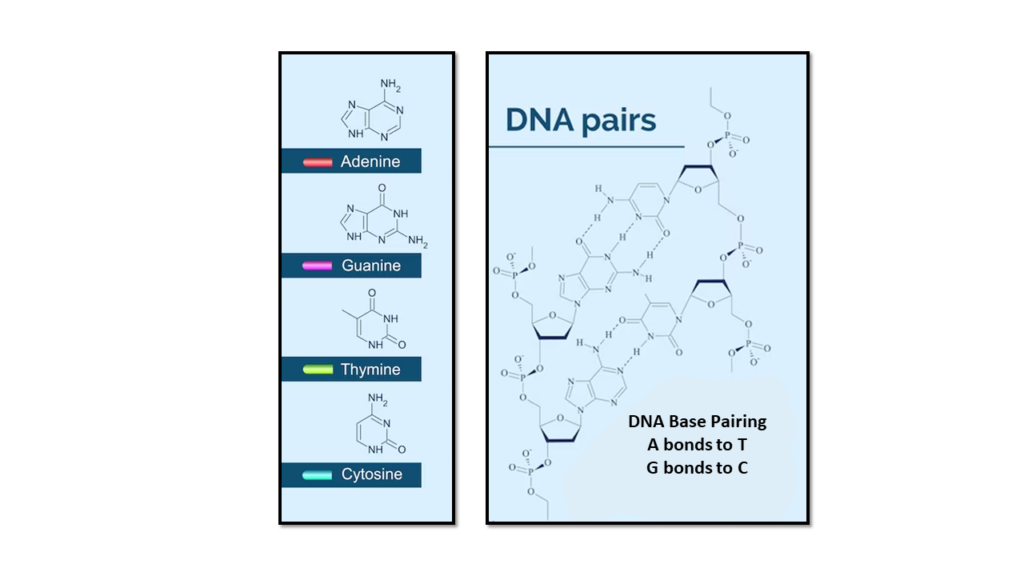
The two DNA strands are known as polynucleotides , composed of simpler monomer units called nucleotides. Each nucleotide is composed of one of four nucleobases ( cytosine [C] , guanine [G] , adenine [A] , or thymine [T]) , a sugar called deoxyribose, and a phosphate group. The nucleotides are joined in a chain by phosphodiester linkage (covalent bonds) between the sugar of one nucleotide and the phosphate of the next, resulting in an alternating sugar-phosphate backbone. The nitrogenous bases of the two separate polynucleotide strands are held together with hydrogen bonds (A with T and C with G) to make double-stranded DNA.
The complementary nitrogenous bases are divided into two groups, pyrimidines, and purines. In DNA, the pyrimidines are thymine and cytosine ; the purines are adenine and guanine .
A large part of DNA (more than 98% for humans) is non-coding, meaning these sections do not serve as patterns for protein sequences. The two strands of DNA run in opposite directions and are thus antiparallel. Both the strands of double-stranded DNA store the same biological information.


DNA isolation and purification are used in laboratories engaged in molecular biology experiments. Several standardized techniques and variations are adapted according to the type of cells or tissues. The process involved lengthy and tiresome ultra-centrifugation in the early days of DNA isolation and purification methods. Now, with the advancement of separation techniques, the procedure is short and agile.
In any method of extraction and purification, there are three main steps: breaking the cells, DNA extraction, and purification.
Cells are broken in different ways depending on the cell type. One standard method for lysis of bacterial cultures is alkaline lysis. In the case of animal cells, lysis is accomplished by detergents or hypotonic solutions.
Plant tissues are homogenized by strong detergents such as SDS (sodium dodecyl sulfate) and heated at high temperatures. Various DNA isolation kits are sold by several biotechnology companies, which are very simple, short, and easy to handle.
The isolation of bacterial plasmid DNA by alkaline lysis method is used for the large-scale isolation of plasmid DNA by modification of the alkaline lysis procedure, followed by purification by phenol-chloroform extraction. Cells containing the desired plasmids are harvested by centrifugation, incubated in lysozyme buffer (re-suspension buffer), and treated with an alkaline detergent. The alkali breaks the cells, releasing DNA and proteins into the medium. Detergent solubilizes the proteins and DNA. The proteins and membranes are precipitated with sodium acetate. The precipitate is centrifuged at a higher RPM, and the supernatant contains the DNA. Finally, the DNA is precipitated by adding 95% ethyl alcohol or propanol. The DNA pellet is re-suspended in a Tris-EDTA (TE) buffer. This DNA sample contains some DNA-binding proteins, which have to be removed. This procedure is often done by phenol-chloroform extraction. This protocol has several variations, each suited to the situation and type of bacterial culture.
Genomic DNA isolation is performed according to the standard protocol suggested by the Federal Bureau of Investigation (USA). The blood samples are stored at -70°C in EDTA vacutainer tubes. Once thawed, a standard citrate buffer is added, the tubes are mixed, then centrifuged. The top portion of the supernatant is discarded, and the additional buffer is added. The tubes are again mixed and centrifuged. The supernatant is discarded, and the pellet is re-suspended in a solution of SDS detergent and proteinase K. This mixture is then incubated at 55°C for one hour. Then the sample is phenol-extracted once with phenol/chloroform/isoamyl alcohol solution and centrifuged. The aqueous layer is removed to a fresh microcentrifuge tube. The DNA is ethanol-precipitated, re-suspended in buffer, and ethanol-precipitated a second time. After the pellet is dried, the buffer is added, and the DNA is re-suspended by incubation at 55°C overnight. A polymerase chain reaction later assays the genomic DNA solution.
Plant tissues bring up several problems during DNA isolation. Plant cells have a rigid cell wall, and the tissue contains many toxic metabolites that can interact with the DNA and change its nature, making it useless for other experimental purposes. Metabolites such as mucilage and other carbohydrates can quickly form complexes with DNA and can damage it. Therefore, the extraction buffer should be supplemented with compounds protecting DNA against these metabolites. Plant molecular biologists widely employ Many DNA isolation techniques using CTAB (Cetyltrimethylammonium Bromide) extraction buffer. This compound forms a complex with DNA and thus protects it from other toxic metabolites such as mucilage and phenolic compounds.
The DNA, isolated and purified by these methods, can be used for various experimental purposes. It can be used for restriction digestion analysis, cloning, ligation, transformation experiments, in vitro transcription, PCR amplification, RFLP (restriction fragment length polymorphism), fingerprinting, RAPID (random amplification polymorphic DNA), sequencing, nick translation and radio labeling, preparation of genomic DNA libraries and cDNA libraries, etc.
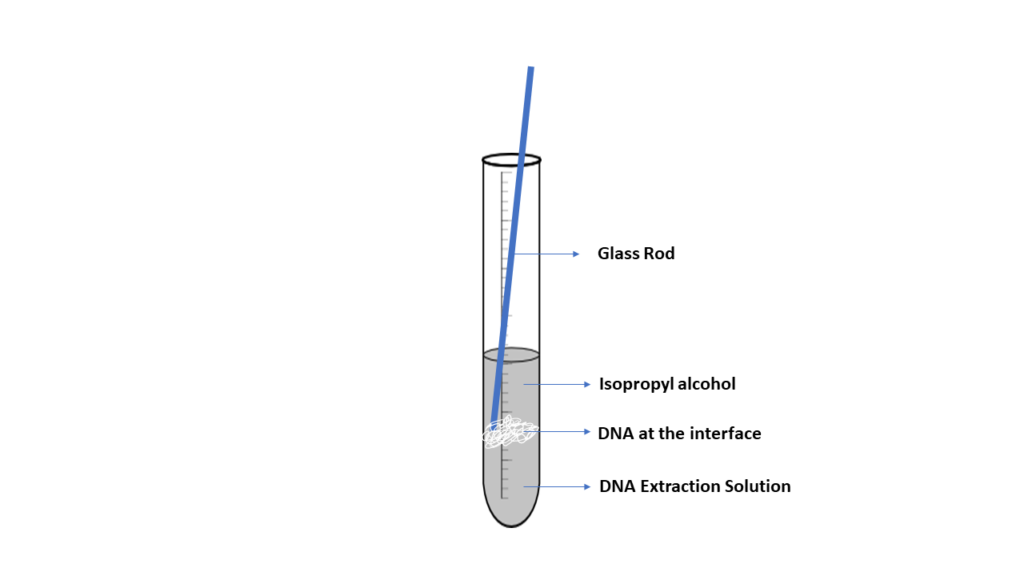
DNA is spooled together using alcohol, which allows DNA fragments to stick together, producing a blob of DNA. When a small layer of alcohol is added to a solution containing cellular fragments and DNA, it will form an interface where the DNA will precipitate. The DNA can then be captured or spooled onto a wooden stick or glass rod. Although this method is effective, the DNA produced is not pure. Other materials, such as protein and cell fragments, are present in the DNA.